Abstract
Background: The presentation of antigens by Major Histocompatibility Complex Class II (MHC-II) is an essential component of adaptive immune response. By combining whole exome sequencing and tandem mass spectrometry (LC-MS/MS), we recently demonstrated that MHC-II presented immunoglobulin neoantigens are common recognition targets in mantle cell lymphoma (MCL) [Khodadoust et al 2017 Nature]. While patient proteomic data can be difficult to obtain, computational methods can learn from these data to predict cancer neoantigen presentation informing personalized immunotherapeutic strategies across cancers. Unfortunately, current tools for predicting peptide presentation by MHC-II have major limitations due to the complexity of presentation pathways and the promiscuity of binding motifs for MHC-II alleles. We hypothesized that a method trained on naturally presented MHC-II ligandomes integrating both sequence and gene expression features could better predict presentation of tumor neoantigens.
Method: We trained a recurrent neural network (RNN) model on 19 mantle cell lymphoma MHC-II ligandomes (>30,000 sequences) to build MARIA (MHC Analysis with RNN Integrated Architecture). MARIA is a deep learning algorithm that predicts peptide MHC-II presentation probabilities based on peptide sequences, neighboring context in each protein (cleavage signatures), patient MHC alleles, and gene expression levels. We evaluated the performance of MARIA with 10-fold cross-validation and also using held out data from both B-cell lymphoma and melanoma patients.
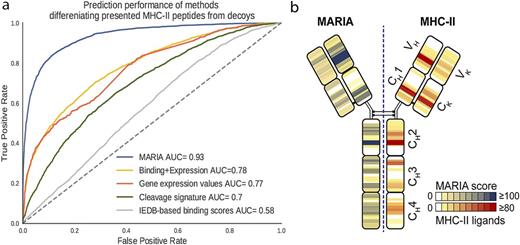
Results: Gene expression levels and cleavage signatures of corresponding peptides have a profound influence on MHC-II peptide presentation but are not incorporated in standard prediction algorithms (Figure 1a). MARIA presentation scores achieved over 0.93 AUC under cross-validation on validated MHC-II ligands from our lymphoma dataset (Figure 1a). In comparison, predicted binding scores alone gave only 0.70 AUC, and conventional shallow neural network models (e.g., NetMHCIIpan3.1) gave 0.87 AUC when trained on the same dataset. When tested on held-out lymphoma and melanoma empirical ligandome data, MARIA sustained over 70% sensitivity with 90% specificity for detection of MHC-II ligands. Though MARIA was exclusively trained on non-immunoglobulin human sequences, it correctly predicted IgM presentation hot spots discovered by direct antigen presentation profiling using LC-MS/MS (Figure 1b), as well as hotspots in alpha-gliadin, a known Celiac Disease antigen, in an HLA-restricted fashion.
Conclusion: MARIA enables high throughput antigen screening with higher accuracy than other methods. It can be applied to immunology applications such as vaccine design, patient profiling, and neo- and auto-antigen identification.
Figure 1. Performance of MARIA predicting human MHC-II peptide presentation. a) Five different predictors of MHC-II peptide presentation were used to differentiate 3290 validation MHC-II peptides from 7500 random human decoy peptides. MARIA scores that incorporate sequence information, gene expression levels, binding scores, and cleavage signatures outperformed other methods with an aggregate AUC=0.93. b) MARIA predicted MHC-II presentation of lymphoma IgM (left) compared to experimentally recovered MHC-II peptides (right). MARIA highlighted MHC-II presentation hot spots on IgM FR3 and CH2 regions, consistent with the experimental heat-map (Spearman R=0.63, p-value<0.0001).
Davis: Vir Biotechnology: Consultancy, Equity Ownership, Honoraria; PACT Bio: Consultancy, Equity Ownership, Honoraria; Adicet Inc: Consultancy, Equity Ownership, Honoraria; Chuga Pharmabody: Consultancy, Honoraria; Amgen: Consultancy, Research Funding; Atreca: Consultancy, Equity Ownership, Honoraria; Juno: Consultancy, Equity Ownership, Honoraria. Altman: Karius: Consultancy; Personalis: Consultancy; Pfizer: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.